PUBLICATION:
SABCS 2020
AUTHORS:
Jennifer A. Crozier, Julie Barone, Kalyan Banda, Beth Lesnikoski, Robert Maganini, Sami Diab, Ian Grady, Thomas Lomis, Charles Cox, Amy M. Truitt, Benjamin Nota, Andrea Menicucci9, Midas Kuilman, Erin Yoder, William Audeh, FLEX Investigators
Background:
Male breast cancer (MaBC) is rare, comprising <1% of all breast cancers in the United States (1). The low incidence of MaBC limits the ability to conduct clinical trials specifically for this population. Due to the paucity of research on MaBC, current understanding regarding MaBC biology, pathology, and treatment strategies has been primarily based on evidence extrapolated from research on female breast cancer (FBC) patients (2,3). Traditional diagnostic biomarkers such as ER, PR, and HER2, as well as newer multi-gene prognostic signatures, are employed when making treatment decisions for both MaBC and FBC (3). However, limited empirical data are available to support the use of identical laboratory biomarkers and molecular signatures in both MaBC and FBC. The 70-gene risk of distant recurrence signature, MammaPrint (MP), and the 80-gene molecular subtyping signature, BluePrint (BP), are commonly used to help make treatment decisions for both MaBC and FBC patients.
To support the clinical utility of MP and BP in MaBC, this study aims to elucidate whether significant molecular biological differences exist between MaBC and FBC. To address this knowledge gap, we evaluated and compared 1) MP index results within Low Risk (LR) and High Risk (HR) groups, 2) MP and BP gene expression, and 3) differentially expressed genes within the full genome and their associated biological pathways between tumors from MaBC and FBC.
Methods
This analysis included a total of 817 breast tumor samples sent to Agendia, Inc.(Irvine, CA) for MP and BP testing. Full-transcriptome microarray data were available on a subset of 480 samples; 1) as previous studies have described the biology of MaBC to be similar to that of postmenopausal FBC (4), a subset from 400 post-menopausal FBC patients enrolled in the FLEX Study (NCT03053193) and 2) 80 MaBC patients, 32 of whom enrolled in the FLEX study and 48 patients for whom limited metadata and quality metrics routinely captured for diagnostic testing were available. Data from all patients were de-identified.
Differences in mean MP indices between FBC (n=400) and MaBC (n=417; 337 MaBC diagnostic samples) according to MP Risk classification (LR or HR) were analyzed using a Z-test. Differential gene expression analysis was performed using the R-limma package in which gene expression data were quantile normalized. Pathway analyses were performed using R-fgsea. Differentially expressed genes (DEGs) were identified between FBC (n=400) and MaBC (n=80) for whom full transcriptome microarray data were available. DEGs were defined as those with a fold change of >2 and an adjusted p < 0.05.
Results
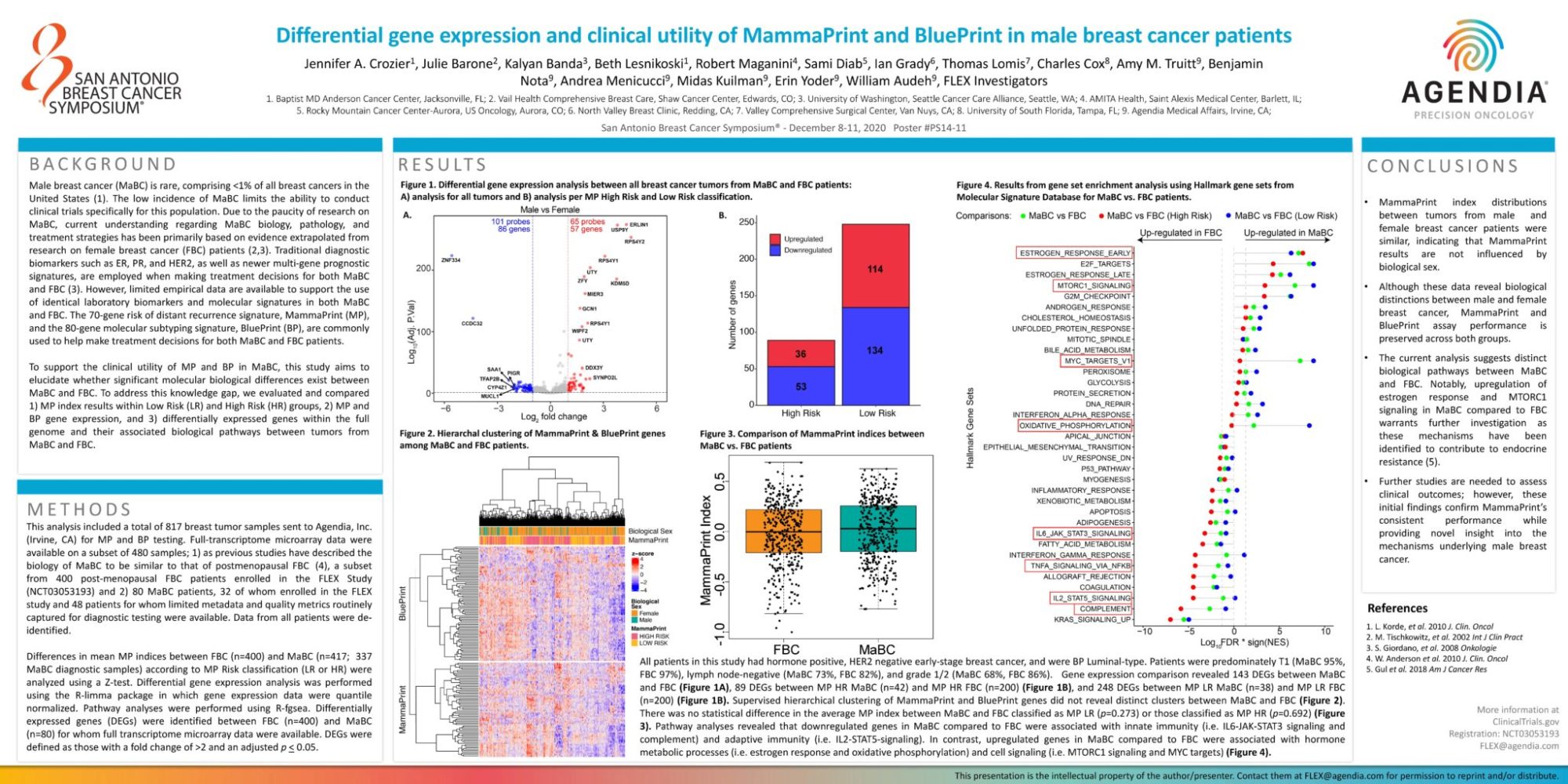
All patients in this study had hormone positive, HER2 negative early-stage breast cancer, and were BP Luminal-type. Patients were predominately T1 (MaBC 95%, FBC 97%), lymph node-negative (MaBC 73%, FBC 82%), and grade 1/2 (MaBC 68%, FBC 86%). Gene expression comparison revealed 143 DEGs between MaBC and FBC (Figure 1A), 89 DEGs between MP HR MaBC (n=42) and MP HR FBC (n=200) (Figure 1B), and 248 DEGs between MP LR MaBC (n=38) and MP LR FBC (n=200) (Figure 1B). Supervised hierarchical clustering of MammaPrint and BluePrint genes did not reveal distinct clusters between MaBC and FBC (Figure 2). There was no statistical difference in the average MP index between MaBC and FBC classified as MP LR (p=0.273) or those classified as MP HR (p=0.692) (Figure 3). Pathway analyses revealed that downregulated genes in MaBC compared to FBC were associated with innate immunity (i.e. IL6-JAK-STAT3 signaling and complement) and adaptive immunity (i.e. IL2-STAT5-signaling). In contrast, upregulated genes in MaBC compared to FBC were associated with hormone metabolic processes (i.e. estrogen response and oxidative phosphorylation) and cell signaling (i.e. MTORC1 signaling and MYC targets) (Figure 4).
Conclusions
- MammaPrint index distributions between tumors from male and female breast cancer patients were similar, indicating that MammaPrint results are not influenced by biological sex.
- Although these data reveal biological distinctions between male and female breast cancer, MammaPrint and BluePrint assay performance is preserved across both groups.
- The current analysis suggests distinct biological pathways between MaBC and FBC. Notably, upregulation of estrogen response and MTORC1 signaling in MaBC compared to FBC warrants further investigation as these mechanisms have been identified to contribute to endocrine resistance (5).
- Further studies are needed to assess clinical outcomes; however, these initial findings confirm MammaPrint’s consistent performance while providing novel insight into the mechanisms underlying male breast cancer.