Source: San Antonio Breast Cancer Symposium®
Authors: Adam Brufsky, VK Gadi, Aisha Ahmed, Preethi John, Beth Seiling, Josien Haan6, Harshini Ramaswamy, Nicole Stivers, Andrea Menicucci, William Audeh, Joyce O’Shaughnessy, FLEX Investigators’ Group
Title: Association of MammaPrint® With Gene Expression Pathways Predictive of Resistance to Cyclin-Dependent Kinase Inhibition
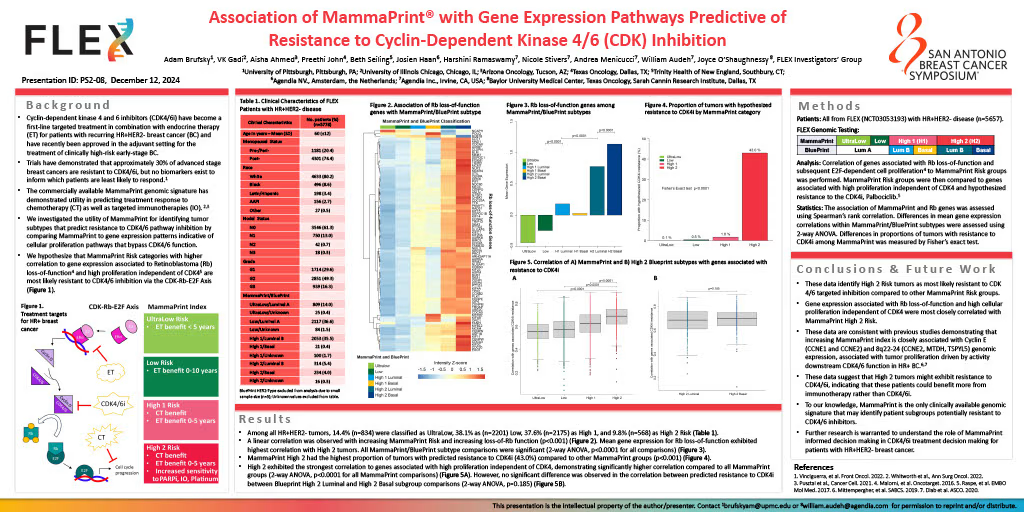
Background: Cyclin-dependent kinase 4 and 6 inhibitors (CDK4/6i) have become a first-line targeted treatment in combination with endocrine therapy for patients with recurring HR+HER2- breast cancer (BC) and have recently been added to the adjuvant setting for the treatment of clinically high-risk early-stage BC. Trials have demonstrated that approximately 30% of advanced stage breast cancers are resistant to CDK4/6i, but no biomarkers exist to inform which patients are least likely to respond. The commercially available MammaPrint genomic signature, characterizing risk of distant recurrence (UltraLow, Low, High 1, and High 2) in early-stage BC, has demonstrated utility in predicting treatment response to chemotherapy as well as targeted immunotherapies. We investigated the utility of MammaPrint in identifying tumor subtypes that predict resistance to CDK4/6 pathway inhibition, using gene expression patterns driven by cellular proliferation pathways that bypass CDK4/6 function.
Methods: All patients with early-stage HR+HER2- tumors enrolled in the ongoing prospective, observational FLEX Trial (NCT03053193), were included in this study (N = 5657). Correlation of ‘Rbsig’ genomic signature, measuring Retinoblastoma (Rb) loss-of-function and subsequent E2F-dependent cell proliferation, to MammaPrint Risk groups was performed. MammaPrint Risk groups were then correlated to an 11-gene signature profile that measures absence of CDK4 phosphorylation, corresponding to Rb loss-of-function and, therefore, predictive of resistance to the CDK4i, Palbociclib. Differences in clinical characteristics were evaluated by Chi-Squared test or Fisher’s exact test. The association of MammaPrint and ‘Rbsig’ was assessed using Spearman’s rank correlation. Differences in 11-gene signature predicted resistance within MammaPrint groups were assessed using Fisher’s exact test.
Results: Patients with HR+HER2- EBC included in this analysis had a mean age of 60 (SD ± 12) and were more likely to be post-menopausal (74.4%). Nearly 31% of patients had T2 stage or larger, 19% of patients had nodal involvement, and 17% had Grade 3 tumors. Among all HR+HER2- tumors, 14.6% were classified as UltraLow, 38.1% Low, 37.4% High 1, and 9.9% High 2 Risk. A linear correlation was observed with increasing MP Risk and increasing ‘Rbsig’ scores (p < 0.001), suggesting MammaPrint High 2 tumors demonstrate highest probability of loss of Rb function. The 11-gene signature comparison demonstrated MammaPrint High 2 as having the highest proportion of tumors predicted to have CDK4 driven loss of Rb function (43.0%), in comparison to UltraLow (0.1%), Low (0.5%), and High 1 (1.8%) tumors (p < 0.001).
Conclusion: These data identify High 2 tumors as least likely to respond to CDK targeted inhibition compared to other MammaPrint Risk groups. The increasing scores of Rb loss-of-function signature, ‘Rbsig,’ was closely correlated with MammaPrint High 2. The 11-gene signature profile, defined by absent CDK phosphorylation and high cellular proliferation, was significantly more likely to be associated with MammaPrint High 2 Risk tumors. These data support previous studies demonstrating that increasing MammaPrint index is closely associated with Cyclin E (CCNE1 and CCNE2) and 8q22-24 (CCNE2, MTDH, TSPYL5) genomic expression, that are hypothesized to identify tumor proliferation driven by activity downstream CDK4/6 function in HR+ BC. To our knowledge, MammaPrint is the only clinically used genomic signature that may identify patient subgroups potentially resistant to CDK4/6 inhibitors. Further research is warranted to distinguish which clinically high-risk, non-High 2 breast cancers are most likely to benefit from CDK4/6i treatments.