PUBLICATION:
AMP 2020
AUTHORS:
Douglas Kingma, Kasey Lawrence, Sahra Uygun, Camila Zanette, Helen Yang, Sammy Mee, Lorenza Mittempergher, Anke Witteveen, Leonie Delahaye, Bob Chan, Annuska Glas
1. Tennessee Oncology, Nashville, TN; 2. Agendia Inc, Irvine, CA; 3. Agendia NV, Amsterdam, The Netherlands
Introduction:
MammaPrint (MP) and BluePrint (BP) are gene expression assays that are utilized in early stage breast cancer (ESBC) patients to aid treatment planning. The MP 70-gene risk of recurrence and BP 80-gene molecular subtyping signatures were developed with microarray (MA) technology and have been commercially available since 2004 and 2010, respectively. In 2018, these tests were translated to Next Generation Sequencing (NGS) using targeted RNA-sequencing and commercialized (with CE mark) for use outside the US1. Equivalence between MA and NGS has been shown previously for MP and BP in centralized and decentralized settings2. Here we present experience with the NGS platform at various global sites and report new equivalence and robustness data for this approach. We also present data for a similar NGS test that has been developed for US use and that is being submitted to FDA clearance.
Methods
Over 1000 samples have been studied with NGS and MA technologies in parallel in Agendia and partner sites. In this substudy, MP and BP results from MA and NGS test were compared for a total of 458 paired samples, 343 from the CE marked NGS kit and 115 from NGS US kit (Table 1). The impact of NGS reagent lots, technicians and testing facilities on the consistency of test results was evaluated. Comparisons of test indices and result categories (e.g., MP Low or High Risk, and BP molecular subtype: Luminal-, HER2-, or Basal-type) were conducted.
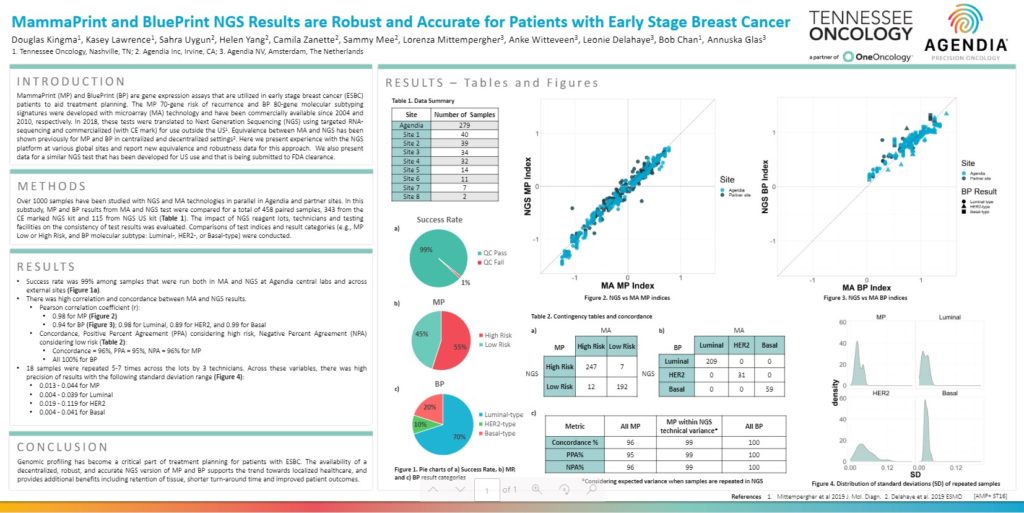
Results
- Success rate was 99% among samples that were run both in MA and NGS at Agendia central labs and across external sites (Figure 1a).
- There was high correlation and concordance between MA and NGS results.
- Pearson correlation coefficient (r):
- 0.98 for MP (Figure 2)
- 0.94 for BP (Figure 3); 0.98 for Luminal, 0.89 for HER2, and 0.99 for Basal
- Concordance, Positive Percent Agreement (PPA) considering high risk, Negative Percent Agreement (NPA) considering low risk (Table 2):
- Concordance = 96%, PPA = 95%, NPA = 96% for MP
- All 100% for BP
- Pearson correlation coefficient (r):
- 18 samples were repeated 5-7 times across the lots by 3 technicians. Across these variables, there was high precision of results with the following standard deviation range (Figure 4):
- 0.013 – 0.044 for MP
- 0.004 – 0.039 for Luminal
- 0.019 – 0.119 for HER2
- 0.004 – 0.041 for Basal
Conclusion
Genomic profiling has become a critical part of treatment planning for patients with ESBC. The availability of a decentralized, robust, and accurate NGS version of MP and BP supports the trend towards localized healthcare, and provides additional benefits including retention of tissue, shorter turn-around time and improved patient outcomes.